Weekly Journal
Week 1
I moved in on Monday, and although it was a bit of a walk, it was nice to find out how lively and convenient the campus is from my dorm. I went to the lab the next day, and I was impressed when seeing the Seibel center for the first time, because the computer science department at my college is much smaller. It was very nice meeting everyone there, but it was a little hectic in the morning because we weren't sure which room had been assigned to us. Over the course of the week, I completed Task 1 of the crash course, and the group held the discussion meeting Friday afternoon. It was my first time reading technical papers, so it was challenging reading through the papers at first, but I was able to understand the material after several read-throughs. It was a good introduction to motion planning, and I learned a lot about the PRM algorithm. I started Task 2 and learned more about the different samplers and the ways in which they build roadmaps. This week, I also started to build my website for the DREU program. It's been a while since I used HTML, so it has been a challenging but fun experience trying to build a website from scratch.
My goal for next Monday is to complete Task 2 and prepare better for the discussion meeting we will have then. I'm looking forward to completing the crash course next week and getting a fuller understanding of the research being done for motion planning. I haven't been able to find a research project that I would like to pursue yet, but I am hoping that, after learning more about motion planning and how it can be applied to different fields like graphics and computational biology, I will have a clearer idea of what I would like to investigate further for the summer. I also plan to increase the functionality of my website in my free time.
Week 2
This week, we completed the Motion Planning Crash Course. For Task 3, the reading article explained more about configuration space, and we got to code our first sampling algorithm that generated random nodes within the configuration space and connected them. For Task 4, we expanded on the code we were given and implemented a simple EST algorithm on our own. This was rewarding for me because it gave me a better sense of the class structures we were using and reinforced the theoretical knowledge that I gained from the algorithms and concepts I read about throughout the crash course. Finally, for Task 5, we had to optimize the EST algorithm that we programmed. For my optimization, I thought that rather than have the start and goal trees grow separately like in the original EST algorithm, it would be more useful for both trees to grow towards the same point in the configuration space at each iteration.
Overall, I think this was a productive week. Because we were able to finish the crash course this week, I'm looking forward to starting research next week. Because we haven't been assigned research tasks yet, I don't have a clear idea as to what I'll be working on next week. However, now that I know more about motion planning, I hope to be able to code more proficiently with the concepts I've learned about in the past two weeks and have a clearer understanding of how to implement the tasks given to us. Another goal will be to continue to read articles and gain more knowledge about motion planning.
Week 3
This week we finally chose our research projects for the summer. I will be working with Diane on her project to use motion planning to compare the accessibility of pathways in bound and unbound proteins. For the project, I will be using the bound and unbound protein structures found in the protein data bank (PDB) to build roadmaps in the free space of the protein and discover tunnels that make the binding sites accessible for the ligands. When setting up the code that I was provided this week, I ran into difficulties running the scripts that would create roadmaps for a given protein and ligand. Because of this, I wasn't able to view the planned path that should be generated with the roadmap of the protein or access the stat file. I started to write the script that would retrieve data like number of nodes and collisions from other stat files that were generated. This will help me get started next week to write the full script to set up the experiment.
For next week, my first goal would be to get all the code to run properly so that I can view the stat file for the path planner of the ligand in the unbound protein environment. My goal for next week is to have written a script that will extract information about the number of paths found and statistics about the energy and clearance levels from the stats files created. The script should organize the data into a more easily readable document that will make it easy to compare the data compiled on the tunnels found for the protein. Once I am able to get the initial experiment to run properly and finish writing a working script, I will try to begin working on getting proteins in bound states from PDB and applying them to the experiment.
Week 4
This week I began searching for matching bound and unbound protein structures to use in the experiment and compile a list of the pairs of proteins in bound and unbound states. While the RSCB PDB contains many PDB files for proteins, there was little documentation on how to find the matching apo (unbound)/holo (bound) structures. Because of this, I had to find a database that could provide such information. I eventually found LigASite which is a database containing matching apo/holo proteins. Although the database has not been updated recently, I think it would provide a good basis for the proteins that I need to use for the experiment.
Next week, I will start by reading the documentation for LigASite to better understand the functions of the database. While the database shows where binding sites are on each protein, because only proteins with internal binding sites are relevant to motion planning problems, I also place to compare the proteins listed in LigASite with another database of a list of known buried site proteins. From this, I will be able to start compiling a list of proteins to use for the experiment. Next week we will also be having our first group meeting to discuss our summer projects, so I will also be working more on my website in addition to preparing to present.
Week 5
This week I continued writing the script to compile data from the statistics generated from building roadmaps and finding paths from the binding site to the surface of the protein. The script generates two tables. One table contains information on the cost and quality of planning the roadmap, including number of nodes, edges, collision detections and run time for each seed. The second table compiles information about the energy, weight and clearance in the tunnels found in each generated seed. Currently, the script can compile data tables for all proteins of a single site (parent folder), and as I begin to build more protein structures, I will try to optimize it further to be able to build data tables for multiple different protein sites at once. I also started to research more on potential protein structures that can be used for the experiment. I had trouble at first building new environments due to the unbound protein structures not containing labelled binding sites. With Diane's help, I was able to solve part of the issue, and it was interesting to look more into the pdb files of the proteins and observe how the two states of the protein shared the same sequence of amino acids but had changed in structure.
I am still running into issues trying to build a new environment, so for next week I plan to start by looking into the issue further by checking if the downloaded files were formatted correctly and if the generated environments and robots contain the correct information. Once I am able to build a new environment properly for the downloaded protein, I will continue to research unbound and bound protein structures and build a dataset of protein structures to use for the experiment. While thinking about how to organize the data I am collecting, I will also the script to factor in changes being made to analyze path similarity and how to optimally present all the data that will be generated.
Week 6
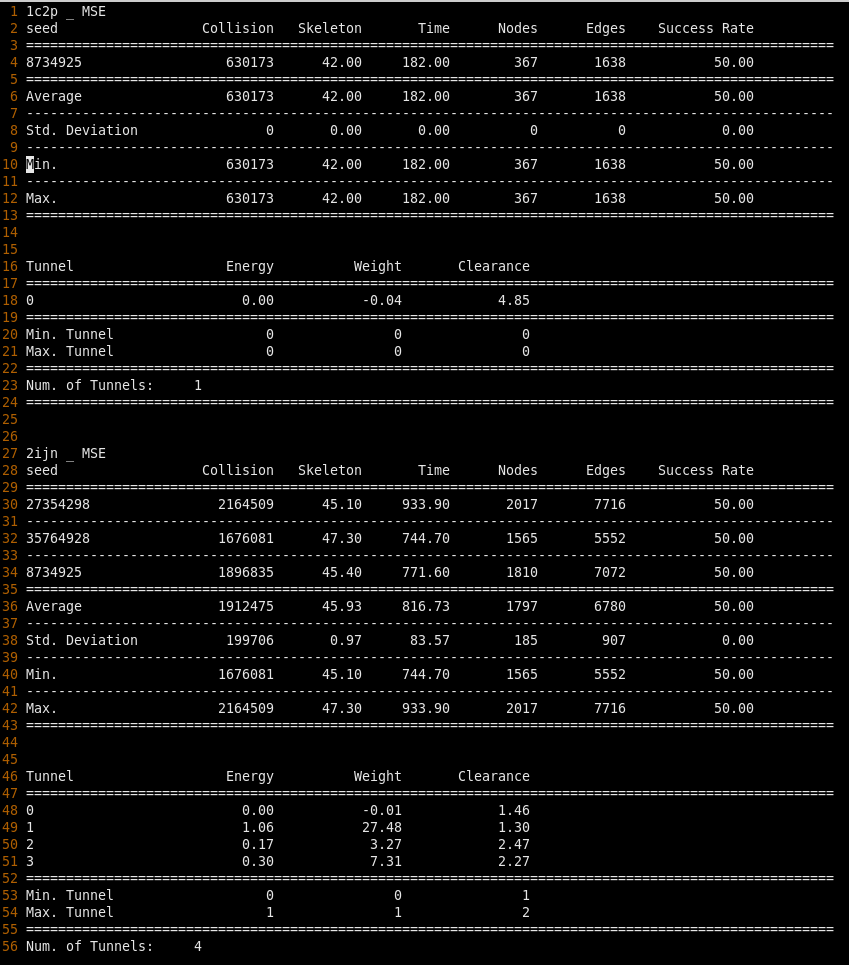 This week, I began to build my dataset of protein structures. The issue I ran into last week seemed to be caused by the generated obj file of the protein structure not overlapping with the pdb coordinates that were provided, causing the planning algorithm to try building a roadmap that didn't correspond to the environment. I looked into the list of buried site proteins that Diane provided me and looked for their corresponding protein structures in the RCSB database. This was difficult because the database doesn't always have apo/holo protein structure pairs. Using LigASite, I was only able to find one protein structure with a holo protein structure pair, so I decided to focus on building an environment with this protein first. With Diane's help, I was able to modify the pdb file by making sure they only contained the active binding site and one protein chain. I ran the algorithm on the holo protein structure (2IJN) first. The script took several hours to finish running several seeds, and from the generated paths, not every seed seemed to be able to find a path, which is something I would like to investigate further next week. However, I was able to run the algorithm with one seed on the apo protein structure beforehand, and when comparing the cost and quality of constructing the roadmap before both proteins, the data suggests a significant
increase in cost in building a roadmap from the apo structure to the holo structure.
This week, I began to build my dataset of protein structures. The issue I ran into last week seemed to be caused by the generated obj file of the protein structure not overlapping with the pdb coordinates that were provided, causing the planning algorithm to try building a roadmap that didn't correspond to the environment. I looked into the list of buried site proteins that Diane provided me and looked for their corresponding protein structures in the RCSB database. This was difficult because the database doesn't always have apo/holo protein structure pairs. Using LigASite, I was only able to find one protein structure with a holo protein structure pair, so I decided to focus on building an environment with this protein first. With Diane's help, I was able to modify the pdb file by making sure they only contained the active binding site and one protein chain. I ran the algorithm on the holo protein structure (2IJN) first. The script took several hours to finish running several seeds, and from the generated paths, not every seed seemed to be able to find a path, which is something I would like to investigate further next week. However, I was able to run the algorithm with one seed on the apo protein structure beforehand, and when comparing the cost and quality of constructing the roadmap before both proteins, the data suggests a significant
increase in cost in building a roadmap from the apo structure to the holo structure.
I will be running the algorithm on the apo protein structure over the weekend so that I can parse the data from the stat files next Monday. My goal for the next week is to continue to research interesting protein structure samples and identify several more protein pairs that will be interesting motion planning problems to use as environments. Now that I have a better sense of how long it might take to run the algorithm and of what to look out for in pdb files before trying to use to them to build environments, I think I will be able to run the experiments more efficiently. I plan to have at least three or four working protein pairs as samples so that I can gather more data and think about how to analyze and discuss the results found.
Week 7
This week I was able to run the experiment successfully on two sets of proteins. This was accomplished with Diane's help in solving various issues with the code, including the issue of generating bad roots that made it hard for the ligand robot to progress through the environment. I also started working on a function that will score tunnels and proteins to judge their accessibility and provide us with a way to quantify our results with a standard scoring system. The score for the tunnels will incorporate the percentage of seeds that were able to discover this tunnel and the min/max energy values that were found in the tunnel. These values each take up a percentage of the score. The score for the protein is similarly structured to incorporate the number of tunnels found and the min/max energy values of the protein. I am still working on what is the best way to quantify the score and will continue to work on incorporating the function next week.
An issue I would like to look into more next week is the Path Analysis across multiple seeds. So far, whenever the algorithm is run, every found best path is returned as its own unique tunnel. However, looking at the tunnels in vizmo shows that many of the tunnels appear to overlap despite the function returning low path similarity scores. Because I have also been working in the PathAnalysis file, I first plan to check that my code has not interfered with the preexisting code before looking into the issue further.
With three weeks left, I would like to start thinking about writing my final report. I have never written a research paper before so I am worried but looking forward to writing one. Although the results I have are still limited, I would like to have most of my first draft done as a goal for next week.
Week 8
This week I continued working on the scoring function. The tunnel score is calculated locally, so it considers the number of seeds that discovered a certain tunnel in comparison to others, the minimum energy in comparison to the average minimum energy across tunnels, and the maximum energy over a max energy threshold (it is set to 1000 for now) that subtracts more from the score the closer the max. energy is to the threshold. The protein score considers the number of tunnels found, the best tunnel score within the protein, and the min/max energy within the protein.
I also tried using CAVER, which is a software tool that analyzes and calculates access tunnels in a provided protein structure. A convenient feature of this software is that it can automatically load given pdb files. I will try to read more about CAVER to understand how the tunnel computation works so that I can use CAVER to generate data that we can use to compare against our own results.
Next week, I will work on writing my final paper and creating a poster. I attended a seminar this week on how to create a good poster, which I thought was helpful. I hope I will be able to apply what I learned to making a poster and be able to finish my first draft next week.
Week 9
This week, I mostly worked on making my poster for the poster session next Wednesday. I met with Diane, and she helped me figure out what should go on my poster and how to layout what I intend to have presented on the poster. I have also been rerunning the experiments this week after receiving the updated code. However, once everyone's code was merged together, I realized the data script that I wrote, while sufficient for the data I need for my poster and paper, might not be fully functional for others because it doesn't take into account some of the new features implemented by the code, so I will update my script to incorporate the new changes.
On Friday, we presented out posters to the lab and got feedback, which was helpful. Because I am still rerunning many of the experiments, which take a long time to complete, I plan to finish filling out the results table Monday once I am able to get the data I need. The main feature I worked on this summer was making a scoring function for the tunnels and proteins. I describe in the Analysis section of Methods and I was wondering if there was a way to present the information more concisely and make it more understandable. I was also unsure of how to layout the Experiments section and if the information I currently included contributes well towards the topic, so I will finish up my poster next Monday after we practice presenting our posters and get more feedback.
I will also work on preparing for the poster session on Wednesday. I have never done one before so I'm not sure what to expect, but I hope that I will be able to prepare what I would like to say well enough to make up for my nervousness about talking and presenting to others. Once I hand in my poster, I will focus on writing up my final paper. Because next week is the last week of the program, I hope to finish the week satisfied with the work I've done this summer and work hard to do well and challenge myself during the poster session.
Week 10
This week was the last week of the DREU program. We had a poster session on Wednesday where we presented the research we did this summer. It was a nerve-wracking experience because we had trouble printing out our posters, but I think the poster session went well and it was a good opportunity to talk with other peers and professors about what I did over the summer and challenge myself with giving presentations. For the rest of the week, I worked on my final paper. Making the poster and presenting in the poster session was helpful in figuring out the outline of what I wanted to talk about in the paper, and with Diane's help, I was able to finish the paper. I also went over the code I wrote to finetune some details and make sure everything was working properly.
It's amazing how fast these ten weeks went by. I learned a lot about motion planning and I think I have a much better idea of what research is like as a student. It was really nice getting to know my peers and mentors and also getting the chance to spend time together outside of lab as well. I am really grateful for having this opportunity to do research at the University of Illinois at Urbana-Champaign with Dr. Amato's lab this summer. It was a new and rewarding experience working with my mentor on a research project, and I am planning to continue to work with my mentor during the semester on this project.