Dong-Hui Xu
DMP Experience 2004
Final Report
Dong-Hui Xu
Motivation:
When I came to realize my interest in applying computer vision techniques to the medical imaging domain, Professor Linda Shapiro recommended me to apply the pixel level texture analysis on one of her on going project "Classification of Protein Crystal Images". This research project is the result of the collaboration between the Department of Computer Science and Engineering and the School of Medicine at University of Washington. The long term goal of the project is to explore the three dimensional structures of key protein molecules and the relationships among protein structure, function and dynamics in order to design new medicine for the treatment of infectious tropical diseases [2]. The short goal is to classify the protein crystal so that the high quality protein crystals could be automatically discovered and further studied of their 3D molecular structures. I was working on the classification goal this past summer.
Background:
Texture is one of the most used features in medical image interpretation, and is applicable to a wide variety of image processing problems. It is often used as a region descriptor in image analysis and computer vision [2]. Texture is a measure of the variation of the intensity of a surface, quantifying properties such as smoothness, coarseness, and regularity. Various methods have been applied towards the analysis and characterization of texture including co-occurrence matrices, discrete wavelet transform, etc.
Although texture is a well-studied property, most work extract texture features either on a global scale or on a uniform texture scale across the whole image. Since texture is a local neighborhood property, texture features computed at a wrong scale would lead to confusion. During the summer, I implemented the approach proposed by Carson et al. [3] for scale selection; the scale selection approach is based on edge polarity, a local image property derived from gradient of L* component of a color image in L*a*b* format, and extracted three texture features (polarity, anisotropy, local contrast) for each pixel on the selected texture scale. These features can be used further in object segmentation and classification.
Approach and results:
While Carson et al. focus on applying the pixel level texture at different scales for outdoor scenes, my main goal during the summer was to apply the approach to medical images, in particular to various protein crystal images.
First, I extracted the three texture features mentioned in the previous section at a scale based on the derivative of polarity measure. Next, with the goal of performing image classification., I used the pixel level texture features to cluster pixels in these images based on their texture similarities. Some of the preliminary results are listed here.
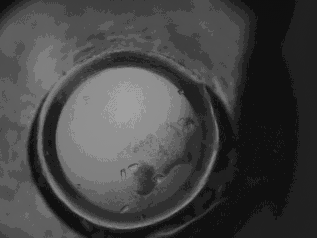
original protein crystal image in JPEG format
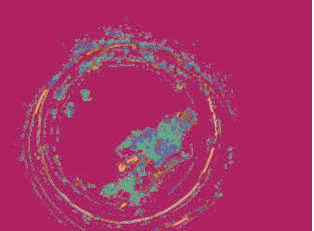
K-mean clustering result (number of cluster is equal
to 10 and similarity measure is Euclidean distance)
Future work:
The three features still needs testing on more medical images in order to see if they have enough discriminatory power to differentiate among different crystal images. I am planning to incorporate more texture models such as Gabor filters and co-occurrence matrix, into the texture analysis as well. Furthermore, I will be looking at some other techniques to derive what is the best scale for the classification in the case of crystal images.
References:
[1] http://www.hhmi.org/research/investigators/hol.html
[2] Chabat, Francis, Guang-Zhong Yang, and David M. Hansell. "Obstructive Lung Diseases: Texture Classification for Differentiation at CT1." Chabat et al. Radiology 2003; Vol 228. pp. 871-877.
[3] Chad Carson, Serge Belongie, Hayit Greenspan, and Jitendra Malik. "Blobworld: Image Segmentation Using Expectation-Maximization and Its Application to Image Querying." IEEE Transactions on Pattern Analysis and Machine Intelligence 2002; Vol 24. pp. 1026-38.