KNN was first introduced by the researchers E. Fix and J. Hodges in their paper, Discriminatory Analysis: Nonparametric Discrimination: Consistency Properties, in 1951.
A distance is assigned between all points in a dataset. Distance is defined as the Euclidean distance between two points or:
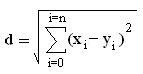
From these distances, a distance matrix is constructed between all possible pairings of points (x, y).
Each data point within the data set has a class label in the set, C={c1,...,cn}.
The data points', k-closest neighbors (k being the number of neighbors) are then found by analyzing the distance matrix. The k-closest data points are then analyzed to determine which class label is the most common amoung the set. The most common class label is then assigned to the data point being analyzed.
In the case where two or more class labels occur an equal number of times for a specific data point within the dataset, the KNN test is run on K-1 (one less neighbor) of the data point in question. This is a recursive process. If there is again a tie between classes, KNN is run on K-2. This continues in the instance of a tie until K=1. When K=1 there is only one class represented in the outcome and thus there can be no tie.
These resulting class labels are used to classify each data point in the data set.
|