 |
Summer Research Project | 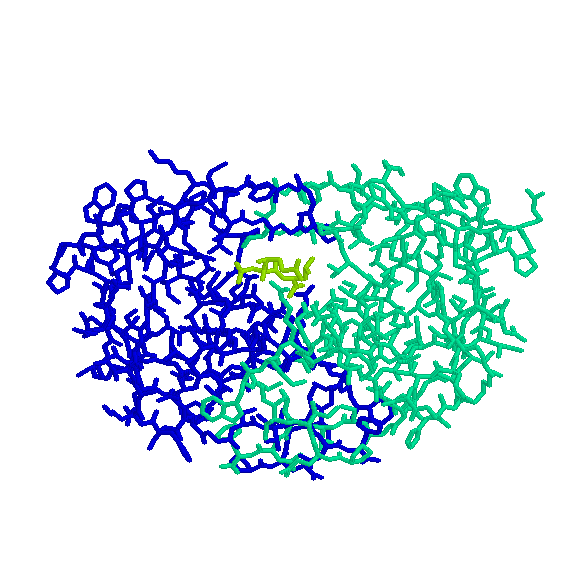 |
 |
Summer Research Project |  |
This summer, I will be working with Dr. Kavraki on a project in the field of Bioinformatics. We will be investigating how to model interactions between large molecules (like protiens, DNA, RNA, etc). We will do this by applying motion planning algorithms (mainly PRMs) to search for valid paths between start and goal conformations of the proteins.
The first step is to understand how to compute molecule conformations (or configurations). In other words, given a set of torsion angles, what are the Cartesian coordinates for every atom in the molecule? To do this, we will first implement a straight-forward approach, the Simple Rotations Scheme, and test it on several small molecules. Then we will look at a more efficient method, the Group Frames Scheme.
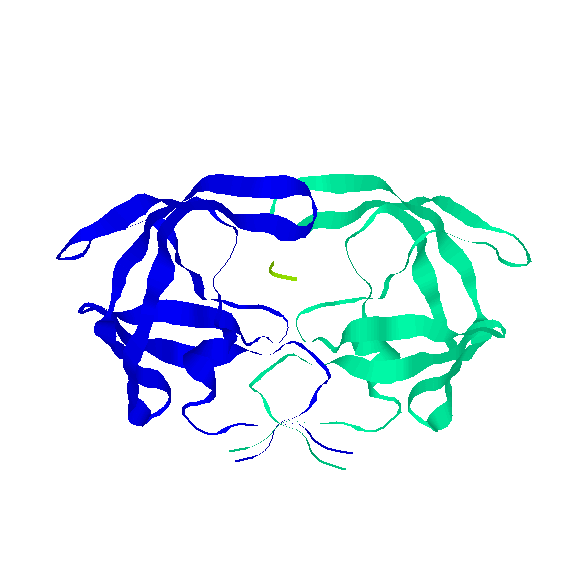
The second step is to perform a protein-protein interaction investigation. We will look for good examples of these interactions, how they function, what their purpose is, etc. and select one to study more in depth.
Once we have targeted a specific interaction, we will develope an algorithm to study the interaction, dynamically. This involves applying PRM methods to the protein and searching for valid paths. Here we have to consider the protein's energy function so the simulations will be realistic.
Weekly Research Journal Final Report
Some useful links for the project:
| Protein Data Bank (PDB) | www.rcsb.org/pdb/ |
| RasMol (A free vizualization tool) | www.umass.edu/microbio/rasmol/ |
Protein-protein interactions we have been studying:
Last updated: 7/16/01